Characterization of the RNase Rae1 in Bacillus subtilis
.
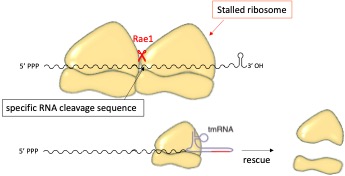
Translation influences the stability of bacterial messenger RNAs. Ribosomes can hinder RNase action by masking endoribonucleolytic cleavage sites or blocking 5'-3' progression. Our project focuses on the Rae1 endoribonuclease from Bacillus subtilis, which associates in a novel way with the ribosome and cleaves mRNAs as they are translated. We identified three Rae1 targets and localized the cleavage sites. Our results suggest that cleavage by Rae1 requires two elements: a cleavage site and a ribosome break (fig1). Our results indicate that, after cleavage, tmRNA enables ribosome recycling. Thus, Rae1 could play a role similar to that of the endoribonuclease CUE2, a player in No-Go Decay in yeast. Rae1 is highly conserved in Firmicutes, Cyanobacteria and plants, where it is localized to the chloroplast. Preliminary transcriptomics experiments have identified potential Rae1 targets in S. aureus and Arabidopsis. Among them, the A. thaliana clpp1 mRNA contains a motif resembling the cleavage site of one of the BsRae1 targets, suggesting mechanistic conservation in plants. The precise mode of recruitment of Rae1 to its cleavage site remains to be elucidated.
Our objectives are to determine (i) which mRNA sequence elements are involved in ribosomal pausing and/or cleavage by Rae1, (ii) how Rae1 interacts with the ribosome and (iii) what role Rae1 plays under physiological conditions that induce translational stress. We plan to elucidate the precise molecular mechanism of Rae1 recruitment to the ribosome, leading to ribosome rescue and destruction of non-functional mRNA in two models of Gram-positive bacteria, and to determine whether these properties are conserved from prokaryotes to eukaryotes.